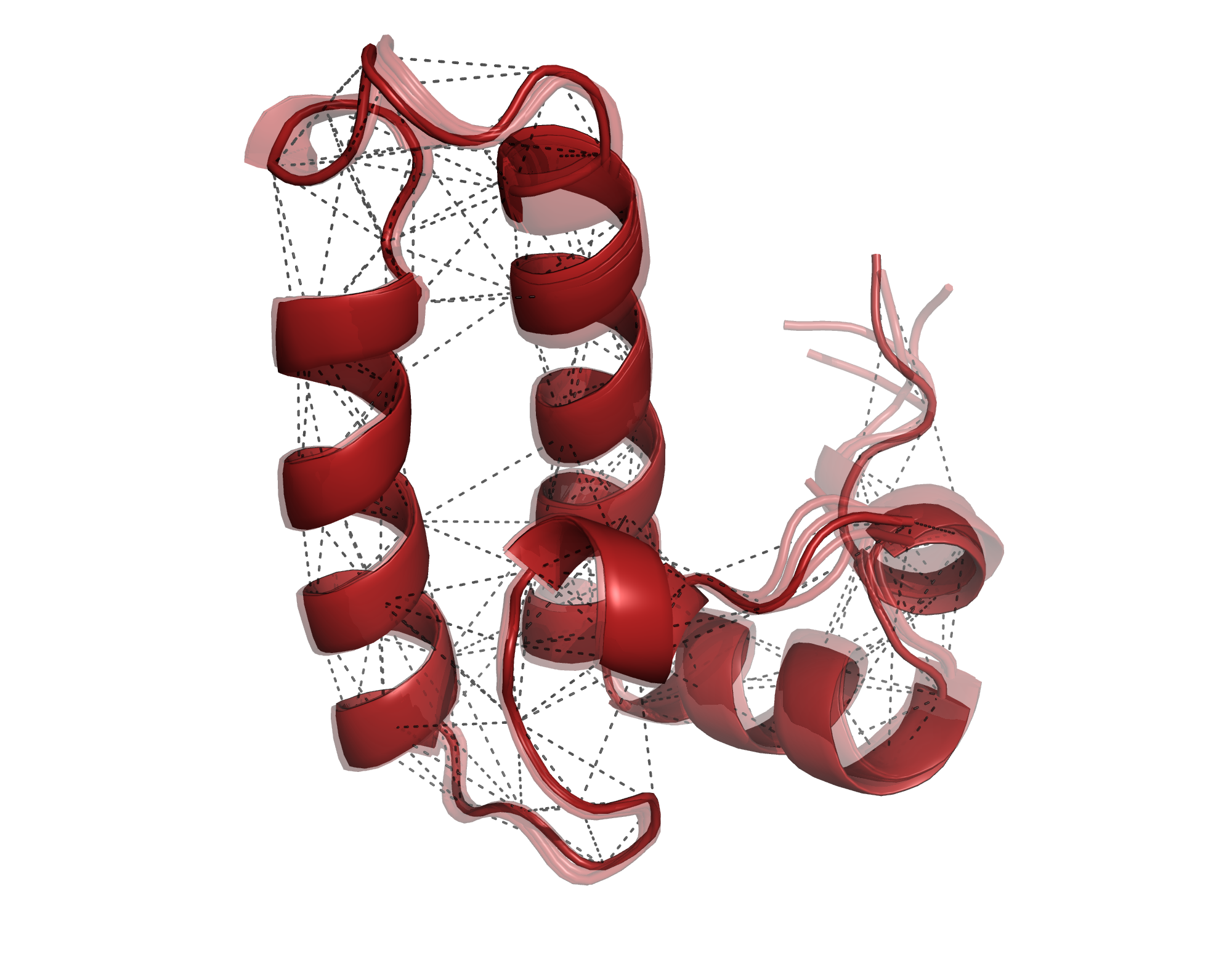
Structure determination
We determine the three-dimensional structure of soluble proteins up to a size of at least 20 kDa in fully automated fashion. An initial sample characterisation will be done to determine the feasibility. Result are delivered within 2 weeks.
Deliverables:
- Structural coordinates (PDB file)
- Report of structure calculation statistics
- Sequence-specific assignment
Sample requirements:
- protein size < 20 kDa
- protein concentration: > 500 uM or > 100 uM in favorable cases
- isotope labelling: 15N, 13C, 1H
- any typical biochemistry buffer (e.g. HEPES, TRIS, phosphate)
- salt concentration: up to 200 mM
Costs:
Academic
- sample check
- up to 20 kDa
free
upon request
Non-Academic
- sample check
- up to 20 kDa
free
upon request
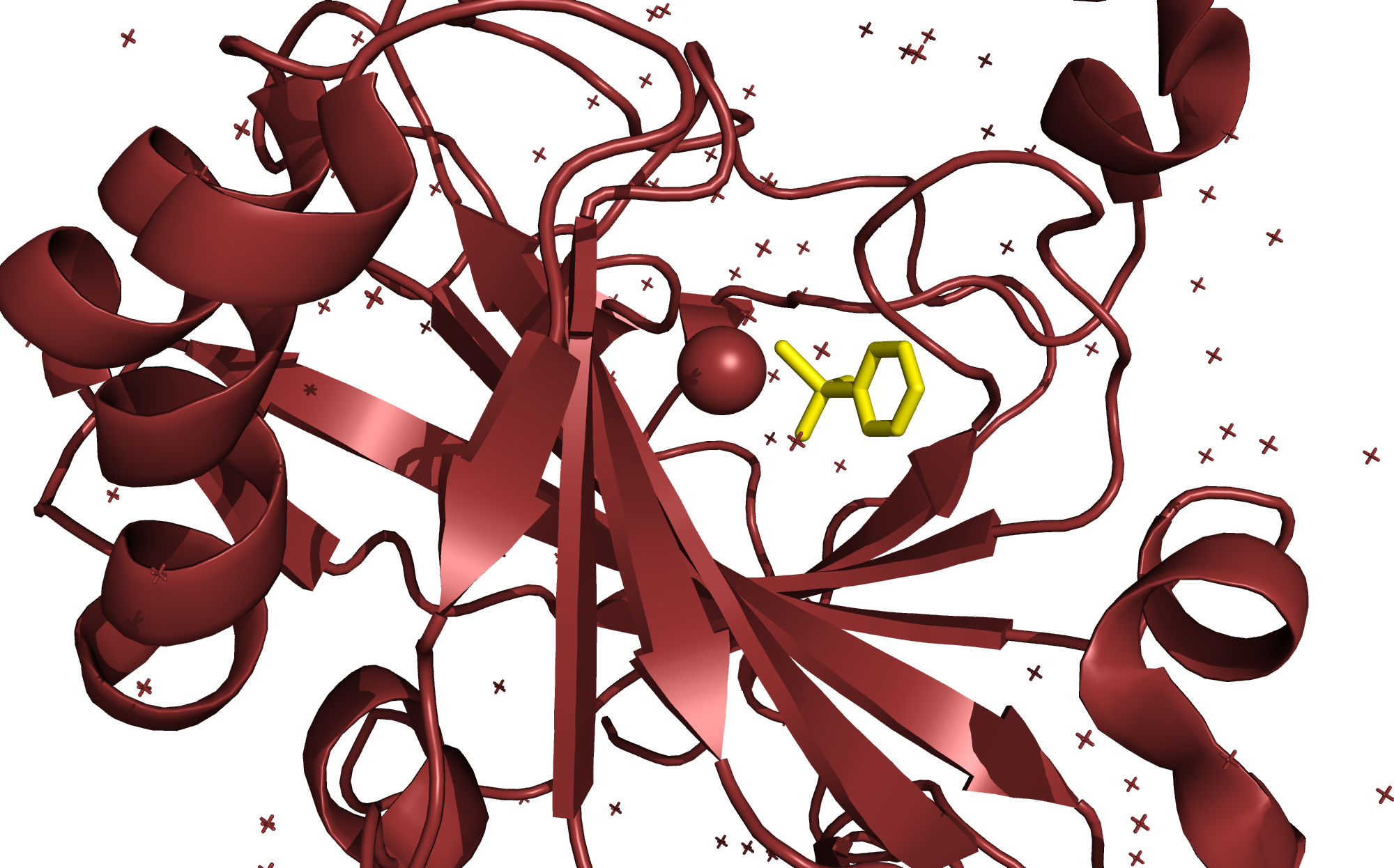
Ligand Binding
We determine the binding site of small molecule ligands on a protein at atomic resolution. Results are delivered within 2 weeks.
Deliverables:
- Chemical shift perturbation data
- Structural visualisation of binding site
- Dissociation constant K D , if in mM to uM range
- Sequence-specific assignment
Sample requirements:
- protein size < 20 kDa
- protein concentration: > 500 uM or > 100 uM in favorable cases
- isotope labelling: 15N, 13C, 1H
- any typical biochemistry buffer (e.g. HEPES, TRIS, phosphate)
- salt concentration: up to 200 mM
- Titratable ligand in separate sample with same buffer
Costs:
Academic
- sample check
- up to 20 kDa
free
upon request
Non-Academic
- sample check
- up to 20 kDa
free
upon request
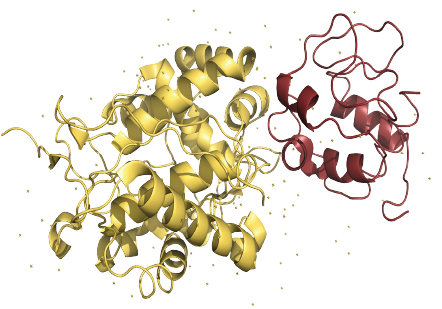
Protein—Protein interactions
We determine the interaction site of proteins at atomic resolution. Results are delivered within 2 weeks.
Deliverables:
- Chemical shift perturbation data
- Structural model of protein-protein complex
- Dissociation constant K D , if in mM to uM range
- Sequence-specific assignment
Sample requirements:
- protein size < 20 kDa
- protein concentration: > 500 uM or > 100 uM in favorable cases
- isotope labelling: 15N, 13C, 1H
- any typical biochemistry buffer (e.g. HEPES, TRIS, phosphate)
- salt concentration: up to 200 mM
- Second protein unlabelled in separate sample with same buffer
Costs:
Academic
- sample check
- up to 20 kDa
free
upon request
Non-Academic
- sample check
- up to 20 kDa
free
upon request

Measurement of protein dynamics
We characterise the backbone dynamics of proteins at the atomic level. Result are delivered within 2 weeks.
Deliverables:
- Spin relaxation data
- Interpretation by model-free analysis
- Structural model of dynamics
- sequence specific assignment
Sample requirements:
- protein size < 20 kDa
- protein concentration: > 500 uM or > 100 uM in favorable cases
- isotope labelling: 15N, 13C, 1H
- any typical biochemistry buffer (e.g. HEPES, TRIS, phosphate)
- salt concentration: up to 200 mM
Costs:
Academic
- sample check
- up to 20 kDa
free
upon request
Non-Academic
- sample check
- up to 20 kDa
free
upon request

Conformational fingerprinting
We characterise the conformational state of a biomolecule in different conditions and compare with reference states. Result are delivered within 1 week.
Deliverables:
- 2D fingerprint spectra
- Quantification of conformational states
Sample requirements:
- protein size < 150 kDa
- protein concentration: > 100 uM
- isotope labelling: selective 13C 1H 3 methyl labelling on 12C, 2H background
- any typical biochemistry buffer (e.g. HEPES, TRIS, phosphate)
- salt concentration: up to 200 mM
Costs:
Academic
- sample check
- up to 150 kDa
free
upon request
Non-Academic
- sample check
- up to 150 kDa
free
upon request

Other Services
Other services can be provided upon request. Please contact us for additional information.
